CLOCS: Contrastive Learning of Cardiac Signals Across Space, Time, and Patients
Within the healthcare industry, the rate of data generation now far exceeds the rate with which such data can be labelled by expert annotators (e.g., medical professionals). The former has primarily been driven by the growing digitization of medical records and the advent of wearable sensors capable of recording health data. The latter can be due to the absence of highly-skilled physicians required for the annotation process (e.g., in low-resource clinical settings) or the disengagement of physicians as a result of being inundated by annotation requests. Irrespective of the cause, the end result remains the same; an environment characterized by abundant unlabelled data and scarce labelled data.
Such an environment would traditionally pose an obstacle to deep learning systems that are notorious for being data-hungry. To overcome this challenge, we propose to exploit the unlabelled data, at scale, such that the deep learning system can achieve clinical tasks better (read: stronger generalization performance) and quicker (read: fewer training epochs) with access to fewer labelled data points. Specifically, we pre-train (read: warm-start) a network by tasking it to solve an arbitrary upstream task. In our case, this involves a framework of contrastive learning that exploits invariances present within cardiac signals. We outline such invariances next.
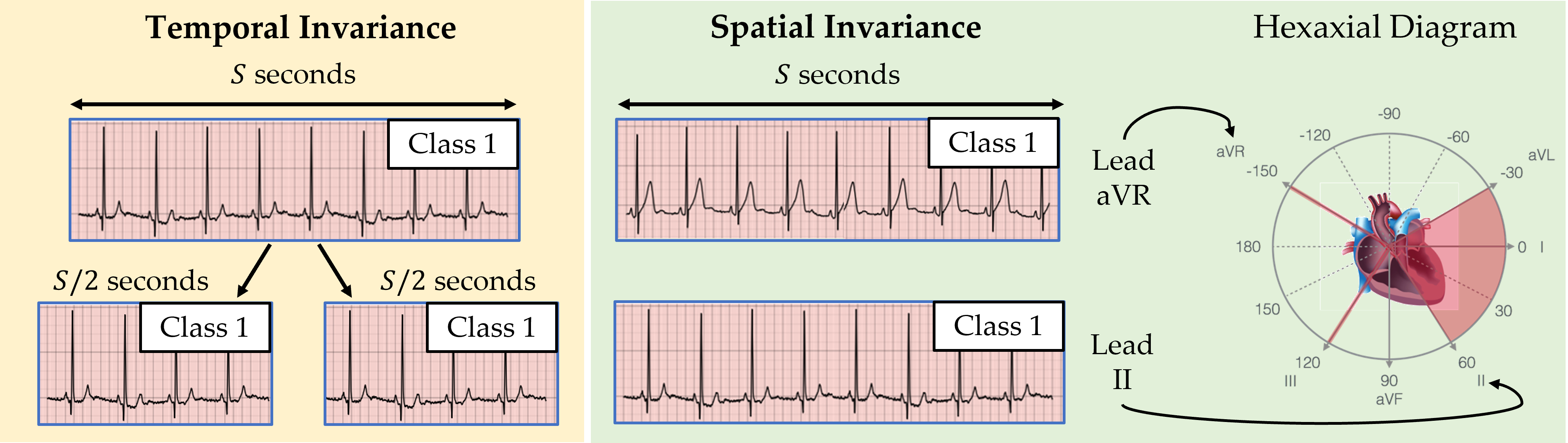
Temporal and Spatial Invariances in Cardiac Signals
At a high-level, an invariance is an aspect of data, which, when changed, does not affect the underlying information reflected by that data. Cardiac signals, for example, can exhibit both temporal and spatial invariances. In this context, temporal invariance (visualized below) implies that temporally adjacent sub-segments of a cardiac signal can be safely assumed to map to the same disease (class). This is a relatively safe assumption to make considering that abrupt changes in the disease are unlikely to occur over a short time-span (e.g., on the order of seconds). For now, we defer an explanation of the spatial invariance of cardiac signals.
Contrastive Multi-Segment Coding (CMSC)
In our ICML 2021 paper, we proposed a family of contrastive learning methods, entitled CLOCS, that exploits spatial, temporal, and intra-patient invariances. A particular member of this family, Contrastive Multi-Segment Coding (CMSC), exploits the aforementioned temporal invariance of cardiac signals as follows (see visualization below). It first retrieves non-overlapping temporally-adjacent sub-segments of the cardiac signal and obtains their corresponding representations (read: vectors that summarize the cardiac signal). We define such representations from the same patient to be a positive pair and attract them to one another. Given a representation of a cardiac signal from a completely different patient, we can form a negative pair of representations which are repelled from one another.
The intuition is that our framework allows for the learning of representations that are invariant to innocuous temporal changes in the cardiac signal.

Cardiac Arrhythmia Diagnosis
Once we complete the pre-training step (i.e., CLOCS), we transfer a subset of the network parameters in order to achieve the downstream task of interest. In our case, this task focuses on identifying the abnormalities in the functioning of the heart, also known as cardiac arrhythmia diagnosis. This involves mapping an electrocardiogram (ECG) signal to a probability distribution over distinct cardiac arrhythmia classes, where each probability indicates the likelihood that the signal belongs to a particular class (see visualization below). A classification can then be made by identifying the class with the highest probability assigned to it.

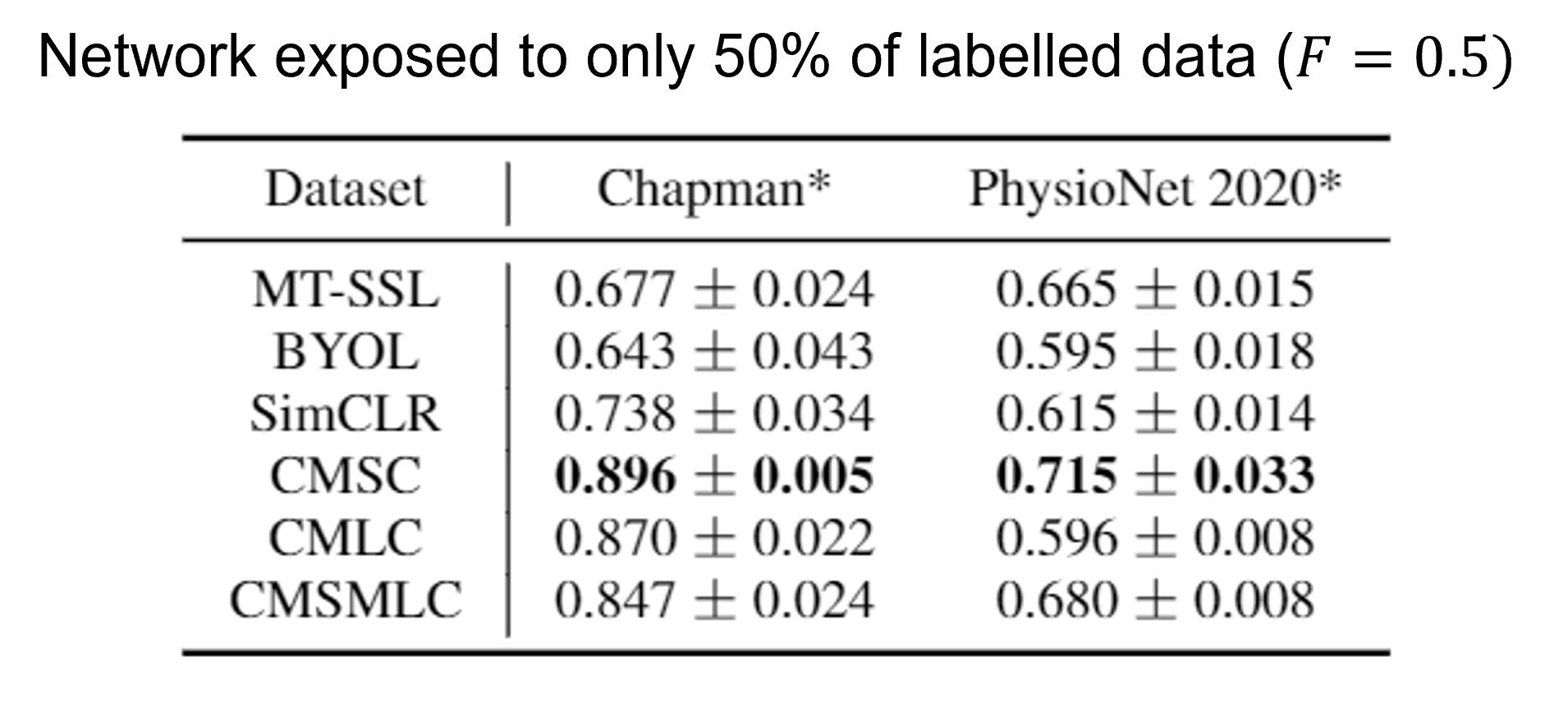
Linear Evaluation of Representations
We deploy our framework in the linear evaluation scenario, where the transferred parameters are frozen (i.e., we extract representations of cardiac signals) and we learn the parameters of a multinomial logistic regression model. Here, we present the results in this scenario where the network is pre-trained on either the Chapman or PhysioNet 2020 dataset and is then fine-tuned on the same dataset while being exposed to only 50% of the labelled data. We show that our framework, CMSC, outperforms both generic and domain-specific self-supervised pre-training methods (e.g., SimCLR, BYOL, and MT-SSL). Such a finding suggests that our framework allows networks to learn rich representations that can transfer well to downstream tasks.
Doing More with Less Labelled Data
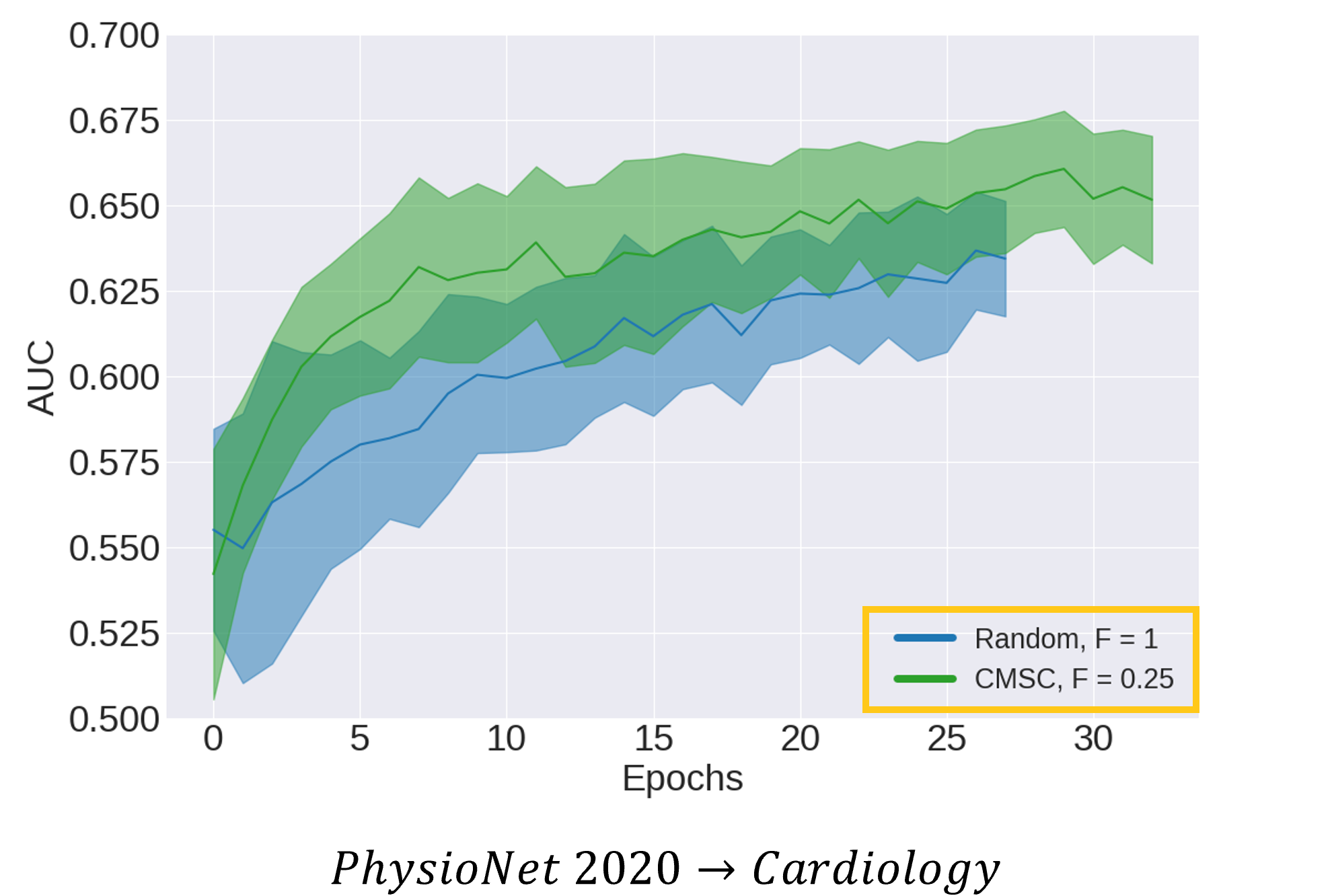
Going further, we wanted to show that CLOCS allows networks to do more (read: learn faster and better) with less (read: fewer labelled data points). To that end, we illustrate the area under the receiver operating characteristic curve (AUC) for data points in the validation set during training for two distinct networks. One network is initialized with random parameters (shown in blue) and is exposed to 100% of the labelled data. The other network is initialized via CMSC (shown in green) and is exposed to only 25% of the labelled data (i.e., a 4x reduction). We show that networks initialized via CMSC learn faster (read: fewer epochs) and better (read: stronger generalization performance) than networks which are initialized randomly, despite a 4-fold reduction in the amount of labelled data. Such a finding exemplifies CLOCS' ability to do more with less labelled data.
Learning Patient-Specific Representations
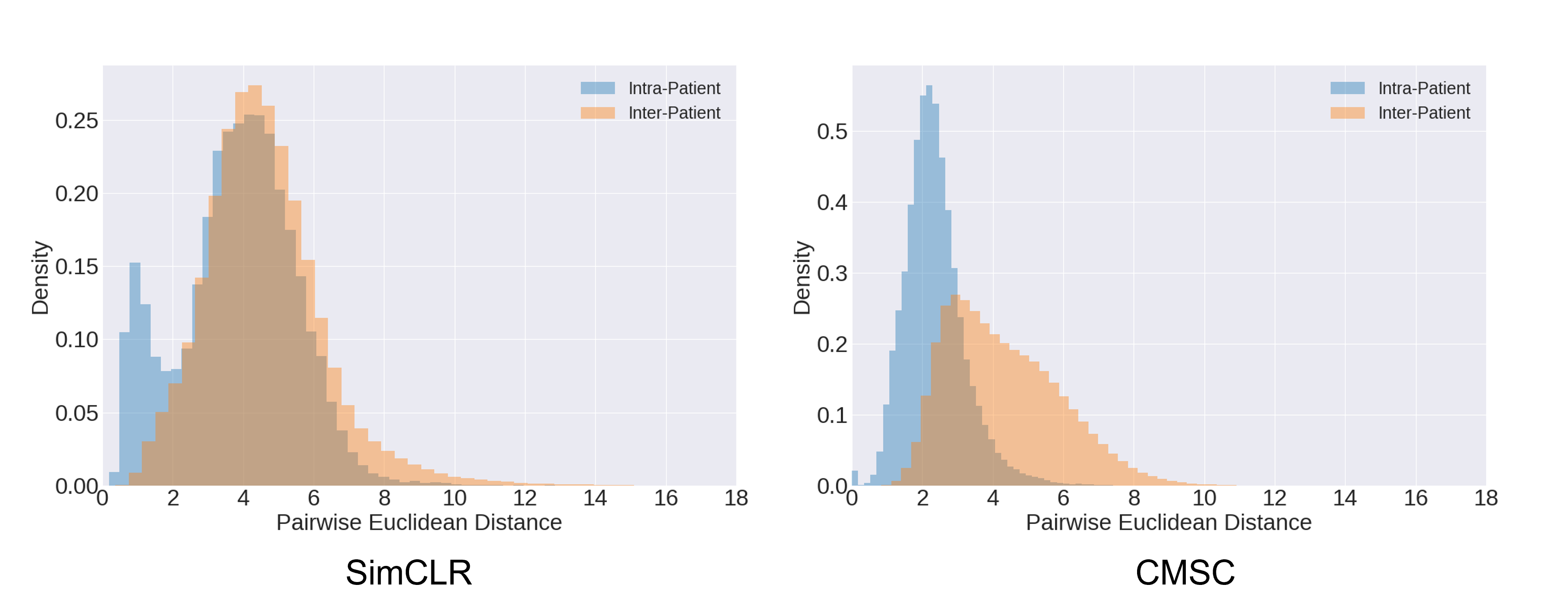
When we plot the distribution of distances between representations of the same patient (Intra-patient) and those between representations of different patients (Inter-patient), we see that the former has a lower mean distance value than the latter. This indicates that CLOCS naturally leads to the learning of patient-specific representations. Such behaviour is absent from other contrastive learning methods, such as SimCLR, as to be expected. We hypothesize that it is this patient-specific behaviour that contributes to the strong performance of CLOCS relative to baseline state-of-the-art methods.
Acknowledgements
We would like to thank Fairuz and Asmahan for lending us their voice.