CROCS: Clustering and Retrieval of Cardiac Signals Based on Patient Disease Class, Sex, and Age
Clinical databases are commonly used by stakeholders within healthcare to search for relevant data points (e.g., medical records) and extract information (e.g., patient attributes) from such data points. For example, cardiologists extract disease information from cardiac signals, researchers search for relevant patients to enroll in a clinical trial, and medical educators search for examplar data points with which to educate the next generation of medical students.
The existing manual search-and-extract process, however, is hampered by two main obstacles; the rapid growth of large-scale clinical databases, and the increased prevalence of unlabelled data points within such databases. Whereas the former implies that the existing manual search-and-extract process scales poorly to large databases, the latter deems the search process intractable to begin with. For example, how would researchers go about searching for relevant patients to include in a clinical trial when such patients are unlabelled with patient attribute information? We propose to address these challenges via the frameworks of clustering and retrieval, as explained next.
Clustering and Retrieval
We aim to extract patient attribute information from unlabelled data points and search for relevant unlabelled data points within a clinical database. To extract information from unlabelled data points, we exploit a supervised clustering approach. In this setting, we learn a set of cluster centroids, each of which is associated with a unique set of patient attributes (e.g., patient disease class, sex, and age). Given an unlabelled data point, we assign it to the cluster centroid to which it is closest, and by extension extract its associated patient attribute information. In essence, we have now annotated this unlabelled data point, thus allowing stakeholders within healthcare to exploit this data point for downstream analyses pertaining to model fairness, error analysis, and beyond.
To retrieve relevant data points from a clinical database, we exploit an information retrieval approach. In this setting, we learn query embeddings (read: vectors), each of which is associated with a unique set of patient attributes. We then use this query to search through the clinical databases, identify unlabelled data points to which it is most similar, and thus retrieve those that are most relevant. This capability allows stakeholders to retrieve patients from clinical databases that otherwise would have been overlooked, a pertinent challenge for clinical trial enrollment. As such, our approach can, for example, allow researchers to recruit a more demographically diverse patient population for clinical trials, thus strengthening the generalizability of the outcomes of such trials.In our research, we focus on explicitly learning the cluster centroids (for the clustering setting) and query embeddings (for the retrieval setting). In practice, these are one and the same, and we will refer to them as clinical prototypes (read: vectors that summarize a unique set of patient attributes).
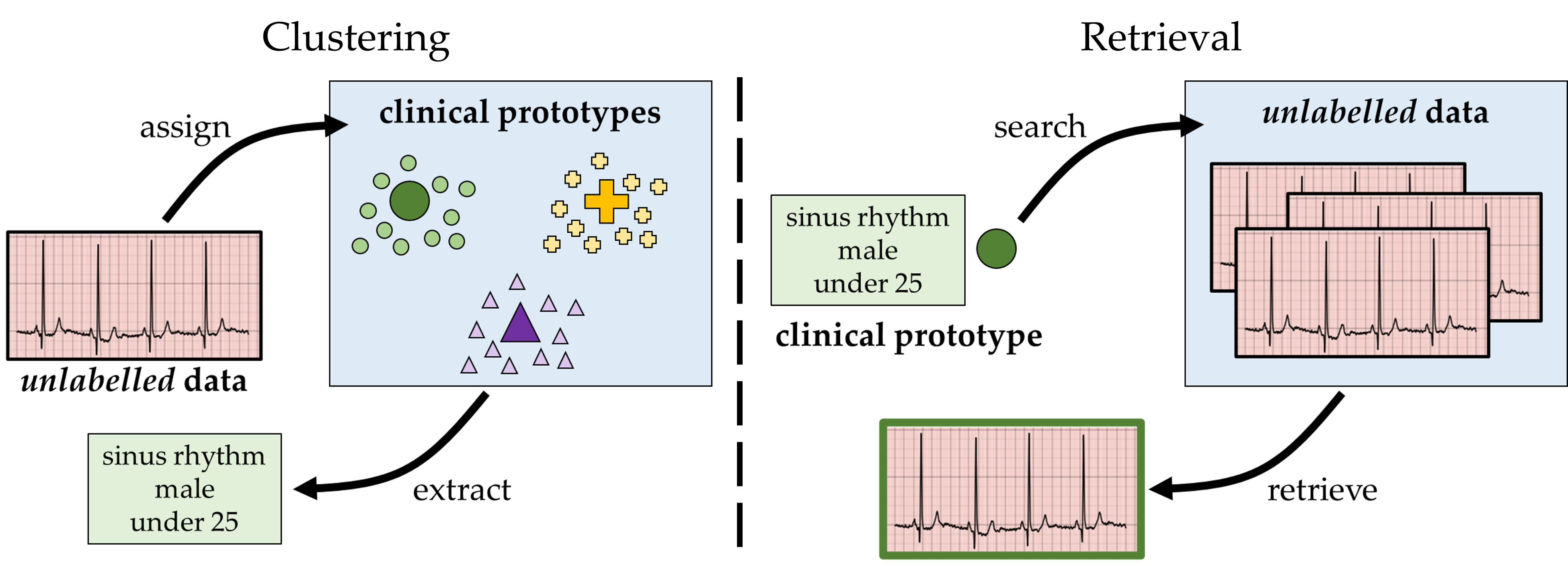
CROCS Framework
In our NeurIPS 2021 paper, we proposed to learn the aforementioned clinical prototypes via a supervised contrastive learning framework, entitled CROCS. To better understand how this works, let us consider the visualization below. For an electrocardiogram (ECG) signal associated with a set of patient attributes, we first extract its representation by passing it through a feature extractor. This representation is then attracted to (or repelled from) the clinical prototypes which are associated with a set of patient attributes. Specifically, it is most strongly attracted to clinical prototypes reflecting the same exact set of patient attributes, and less attracted to those with some overlap in the patient attributes. At the same time, the representation is repelled from clinical prototypes associated with a different disease class.
In doing so, we learn embeddings that are attribute-specific and which can serve as either centroids in the clustering setting or queries in the retrieval setting. Note that clinical prototypes are embeddings that are learned in an end-to-end manner, similar to how network parameters would ordinarily be learned.

Interpretable Clinical Prototypes
We also impose an additional constraint on the clinical prototypes to increase their level of interpretability. More precisely, we encourage prototypes that are associated with a similar set of patient attributes to be more similar to one another. For example, prototypes that share the same disease class and sex group but differ according to the age group should be more similar to one another than those from distinct sex and age groups. Doing so results in the visualization below, where we illustrate a low-dimensional projection of the learned clinical prototypes (marker style = disease class, colour = sex group, marker size = age group). The main takeaway here is that the clinical prototypes adopt a semantically meaningful arrangement, as desired.
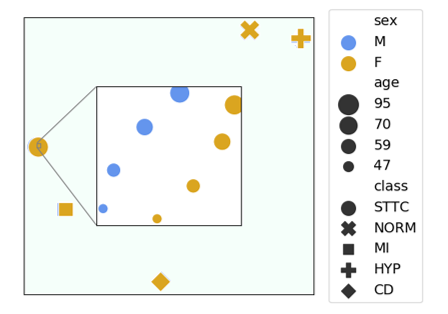
Visualizing Clinical Prototypes
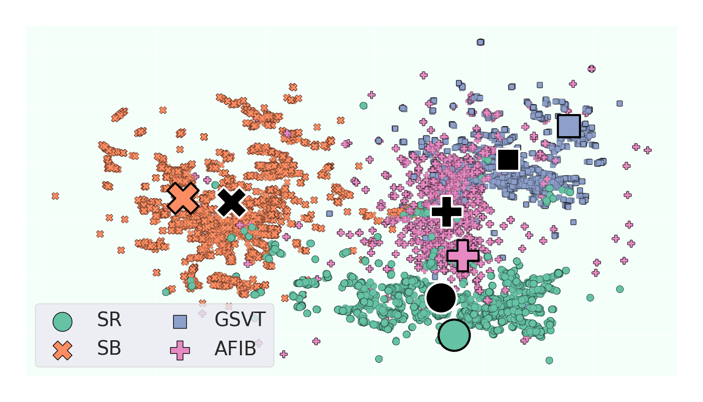
When we project the clinical prototypes (and the data points in an unseen held-out set) onto a low-dimensional space (e.g., via UMAP), we arrive at the visualization below. Representations of instances in the validation set are shown as small, coloured points. Traditional prototypes, which are av average of representations associated with a unique set of patient attributes, are shown as large, black points. Clinical prototypes are shown as large, coloured point. We see that these clinical prototypes (large, coloured points) are disease class-specific and align well with the class labels of the data points in the unseen held-out set. This bodes well for their eventual use in the clustering and retrieval settings. They are also distinct from traditional prototypes (large, black points).
Clustering Results
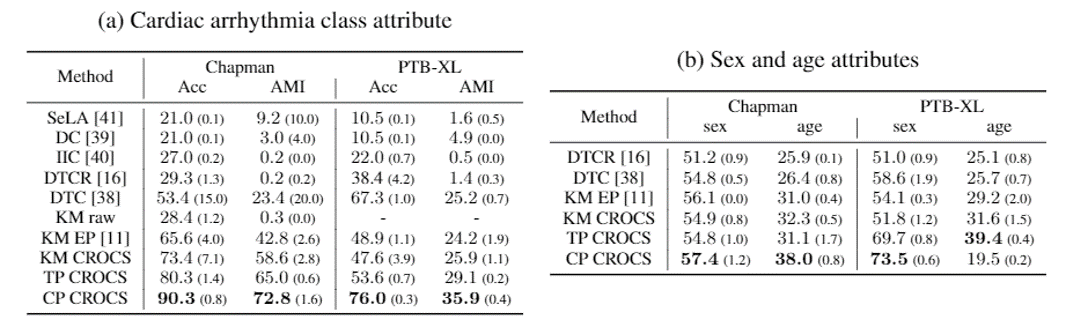
In the clustering setting, we exploited the learned clinical prototypes as centroids of attribute-specific clusters. Here, we compare their performance to that of baseline methods when clustering multiple patient attributes (namely, cardiac arrhythmia disease class, sex, and age). The main takeaways is that CROCS outperforms the state-of-the-art method, DTC, across datasets and attributes. As such, if you are looking to cluster data points according to multiple patient attributes, we recommend using the CROCS framework.
Retrieval Results
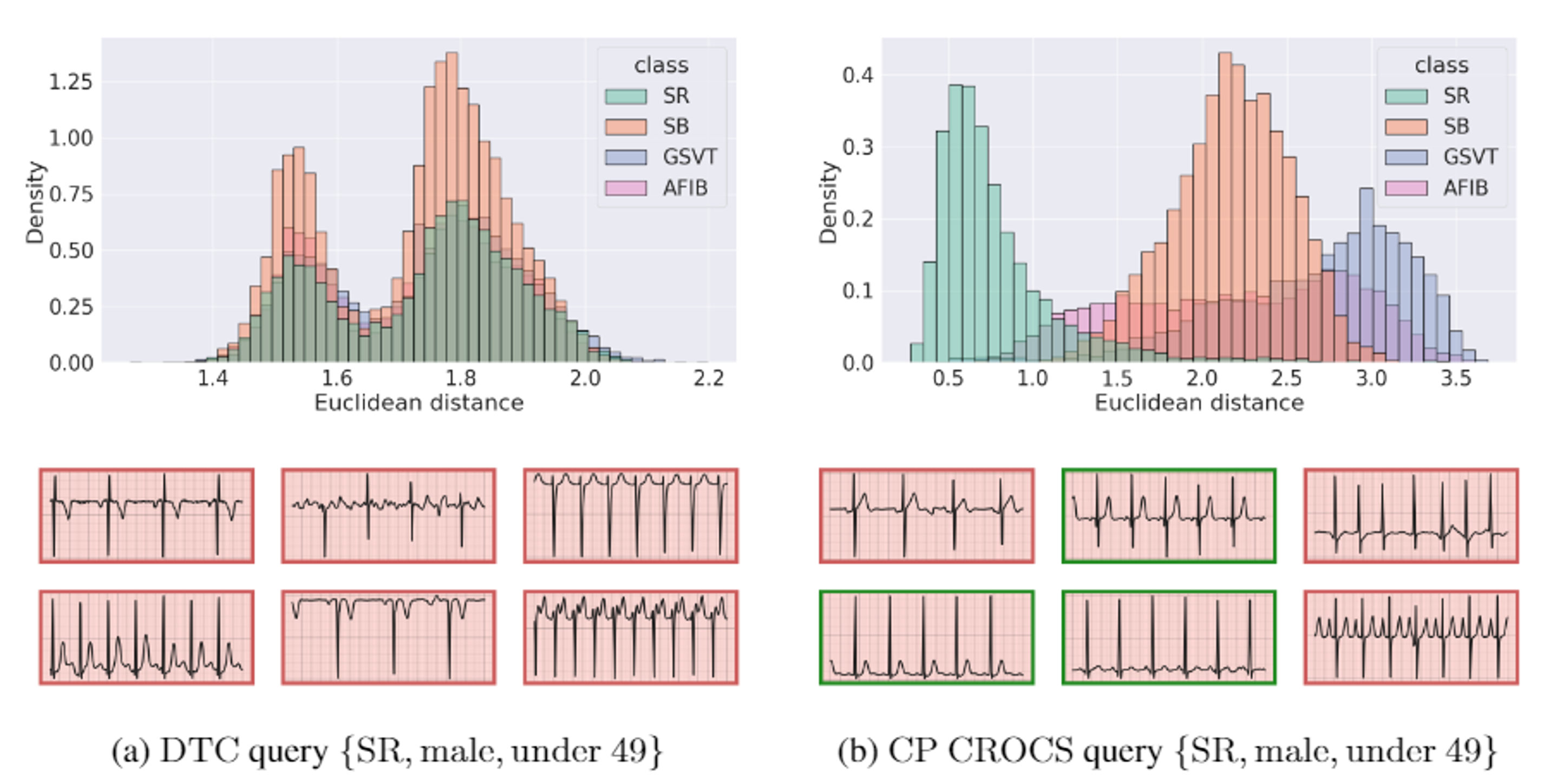
In the retrieval setting, we exploit the learned clinical prototypes as queries to search through a database of unlabelled cardiac signals and retrieve those which are most similar. Doing so for a particular clinical prototype (associated with sinus rhythm disease class, male sex group, and the age group under 25 years old) leads to the results below. We find that this prototype is closer to unseen data points which belong to the same disease class than to those from a different class. This suggests that CROCS is likely to retrieve relevant cardiac signals, a claim supported by the relevance of the six closest ECG signals (green bounding boxes indicate relevant cardiac signals). Such behaviour is in stark contrast to queries learned by DTC, which struggle to retrieve relevant cardiac signals.
Acknowledgements
We would like to thank Antong Chen for reviewing a preliminary version of the associated manuscript. We would also like to thank Nagat Al-Saghira and Mohammed Abdel Wahab for lending us their voice.